Introduction
The last two decades started a change in how microbiological methods are being performed, using rapid microbiological methods (RMMs) and alternative microbiological methods. In many places, microbiological identification systems are treated separately from RMMs. Since the completion of the human microbiome project, a need for new tools has been identified for identifying the specific organisms and traits in the various biome applications.1 While researchers are evaluating the microbiome for various parts of the body, e.g., blood, urinary tract, gut, reproductive tracts, and the like the need for new tools is frequently reported.
In recent years, much of the microbiological development taking place is in the appropriate identification methods to be used. Much of this research has resulted from the sequencing of the entire human genome. Historically, microbiological strains were characterized and defined based upon the available microbial isolates.2 Today, there are newer technologies and methods that have allowed for the assessment of microbial communities in different ways, looking at their ecology and phenotypes within the community and the human microbiome. With a worldwide pandemic going on, there is an increased focus on the potential consequences of small genetic variations in pathogenicity of microbes and its effects on human health.
The newer technologies allow for high-throughput, most often culture-independent techniques. These technologies allow for the identifying, quantifying, and tracking strains via high throughput sequencing supplemented by molecular and “culturomics” methods.3 This article discusses some of the newer methods that are being used.
Hierarchy of Methods from DNA to Phenotype
In assessing tools to use in the microbial identification process, there is an established “Central Dogma of Biology”. This approach is shown in Figure 1. It goes from genomics to transcriptomics, to proteomics, to metabolomics, to biological phenotype. Metagenomics would be in the “genomics” category, but different in that it is the genomics of a group of varying microorganisms. In pharmaceutical applications, we are somewhat familiar with genomic identification and phenotypic identifications. In recent years, mass spectroscopy methods have been implemented for a quick, high throughput method of microbial identification, i.e., proteomics. In many ways, this has revolutionized the ability to process large numbers of environmental isolates at a low cost and in a short time. Other members of this hierarchy will be discussed.
Methods for the Preparation of Clinical Samples
Microbial genomics have advanced significantly using high throughput Whole Genome Sequencing (WGS). WGS is the preferred method for pathogen sub-typing. WGS has been widely used in the study of etiology, evolution, and transmission of various types of diseases, and antibiotic resistance. The inherent problem with current culture based WGS is related to the requirement of generating a laboratory culture of the pathogen to be studied.4
There is a significant benefit in the use of diagnostic tests that do not require the culture of the organism/pathogen. These are designated molecular-based culture independent diagnostics tests (CIDT), especially in the time required for testing and the time to results.
As laboratories have gone to these types of methods, the downside is that it reduces the numbers of isolates available for typing. When using culture independent methods, no isolates are recovered for use in subsequent testing and evaluation. In turn, this can reduce the capacity of surveillance systems to detect clusters and identification of the source of outbreaks. As we utilize more CIDT notifications, alternative methods for identification and characterization of pathogens that can cause potential epidemics need to be identified.5
Metagenomics provides the ability to provide unbiased sequencing of all the DNA in a clinical sample without the cultivation of all the separate isolates. Historically, metagenomics was focused on the make-up of environmental and human ecosystems, especially in understanding the diversity. More recently, metagenomics has been useful in identifying emerging infectious agents. If these methods can be used to sequence directly from clinical specimens, it can improve the surveillance of antibiotic resistance and transmission.6
Metagenomics – Shotgun Sequencing
Metagenomics refers to the study of genetic material recovered directly from environmental samples. Since the genes are studied within a community, it allows for assessing the functional capabilities of a given microbiome. Knowing that a gene can perform a function is not the same as knowing whether it is expressed (i.e., transcribed and translated).8
Shotgun sequencing is often used for performing metagenomics. In shotgun sequencing, the extracted DNA from the community is sheared and sequenced. The results are “blasted” (compared) to different databases (e.g., the functional database: Kyoto Encyclopedia of Genes and Genomes (KEGG) or the protein database: Clusters of Orthologous Groups (COG)) to assign genes or pathways. For example, some genes may code for translation, signal transduction, secondary metabolite biosynthesis, transport, and catabolism.9
Subscribe to our e-Newsletters
Stay up to date with the latest news, articles, and events. Plus, get special offers
from American Pharmaceutical Review – all delivered right to your inbox! Sign up now!
Unless the sequencing is extensive, it can be difficult to determine some of the functions. This is a significant downside of metagenomic analysis. There is no consensus in the literature on just how much sequencing is needed.10
Frequently, the genetic sequences identified may not yet be assigned or they may not yet be found in the available databases.11
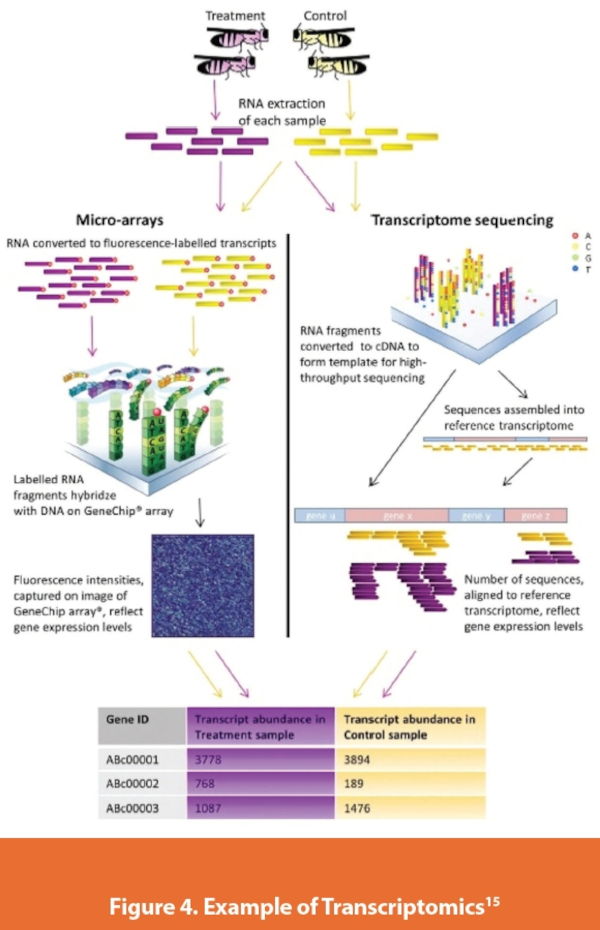
Metatranscriptomics
Metatranscriptomics refers to the study of gene expression in microorganisms in their natural environment. It provides a way to assess whole gene expression profiling of a complex community of microorganisms. It allows for the study of the diversity of the active genes in the community. This technology is like metagenomics. It assesses both the functional potential of a microbial community as well as assessing the genes that have been activated and transcribed versus those that are not. This provides a way to measure the actual metabolic activity of a microbial community.13
The evaluations conducted can be quite complex and there are many unrecognized functional RNA elements. The process starts with the extraction of the total RNA from the community sample, conversion of total RNA, or enriched messenger RNA (mRNA) to complementary DNA, can combine with sequencing to assess the expressed portions of the genome. Often, this process is difficult.14
Metabolomics
Metabolomics refers to the study of the group of metabolites present within an organism, cell, or tissue. It aids in the ability to quantify and identify small molecules, which are metabolism byproducts. Creating the metabolic profile is used like the term microbiota is used to describe all the bacteria in a present environment. The organism’s genetics and environment affect the metabolism. Metabolomics allows one to monitor the outcome. Monitoring this information allows for greater understanding of the function and effect of the metabolite.
The most common ways to assess the metabolic profile are mass spectrometry (MS) and nuclear magnetic resonance (NMR). The technologies work based upon different processing methods. MS requires pairing with an upstream separation technique, e.g., gas chromatography (GC-Moro liquid chromatography (LC-MS). It determines the differences in molecules based upon their mass-to-charge ratios and retention times. NMR, since it is non-destructive, allows for the sample to be recovered and used for additional evaluation. The datasets generated are complex and can be difficult to analyze.
Next-Generation Sequencing (NGS)
Several scientists are using NGS to study microbial communities. The benefit of this technology is bypassing the limitations of current culture methods. This technology is used for the study of genomes or transcriptomes of microbial communities. Unfortunately, it will take more than identifying the strains present in a community to understand the community. It is important to analyze the functions and interactions of the constituents of the community. For example, one needs this type of analysis to understand the biological mechanisms that may explain the relationship between the microbiome and the health of the host. This understanding must be based upon living communities, not the processed library catalogs used for many NGS systems.
Conclusion
It is an exciting time in microbiology, with all the research relative to the microbiome and disease. We need new approaches to culturing microorganisms. Culturing microorganisms is an issue in traditional pharmaceutical microbiology methods also. For example, there are so many issues with the inability to recover organisms in a sterility test method that most times contaminated samples would not be detected. Campylobacter jejuni is a significant pathogen for infants. If it were present in a sterility test sample, it would never be detected because it only grows at a temperature condition outside the limits of the compendial sterility test. Ideally, it would be good to study living organisms and allow for high-throughput analyses as needed in modern science. It would be beneficial to have automated methods to reduce the preparatory work needed, high-capacity analyses, and the ability to process a high number of strains concurrently. Lastly, like all microbiological testing, it needs to be economical.

References
- Christey, P. (issued by Hall, H.) (2020) As microbiome studies surge, new tools are needed. R&D World. Downloaded from: https://www.rdworldonline.com/as-microbiome-studiessurge-new-tools-are-needed/
- Yan, Y., Nguyen, L.H., Franzosa, E.A., and Huttenhower, C. l. (2020) Genome Medicine 12:71 https://doi.org/10.1186/s13073-020-00765-y.
- Yan, Y., Nguyen, L.H., Franzosa, E.A., and Huttenhower, C. l. (2020) Genome Medicine 12:71 https://doi.org/10.1186/s13073-020-00765-y.
- Bachmann, N.L., Rockett, R.J., Timms, V.J. and Sintchenko, V. (2018). Advances in Clinical Sample Preparation for Identifi cation and Characterization of Bacterial Pathogens Using Metagenomics. Downloaded from: https://www.frontiersin.org/articles/10.3389/fpubh.2018.00363/full on October 14, 2020.
- Bachmann, N.L., Rockett, R.J., Timms, V.J. and Sintchenko, V. (2018). Advances in Clinical Sample Preparation for Identifi cation and Characterization of Bacterial Pathogens Using Metagenomics. Downloaded from: https://www.frontiersin.org/articles/10.3389/fpubh.2018.00363/full on October 14, 2020.
- Bachmann, N.L., Rockett, R.J., Timms, V.J. and Sintchenko, V. (2018). Advances in Clinical Sample Preparation for Identification and Characterization of Bacterial Pathogens Using Metagenomics. Downloaded from: https://www.frontiersin.org/articles/10.3389/fpubh.2018.00363/full on October 14, 2020.
- Downloaded from: https://www.bing.com/images/search?view=detailV2&ccid=3yW87V6W&id=DE69AB6EDBD943762C0BAA273B4C33CE86F201C2&thid=OIP.3yW87V6WKQ_egL2Ig7uUtQHaEG&mediaurl=https%3a%2f%2fwww.lcsciences.com%2fdocuments%2fsample_data%2fmetagenomics%2fsrc%2fpics%2fmetagenomics_overview.jpg&exph=479&expw=865&q=image+metagenomics&simid=608007871451106944&ck=033F893FA2D74273EDCA5EFB95485310&selectedIndex=7&FORM=IRPRST&ajaxhist=0.
- Smith, M.I., Turpin, W., Tyler, A.D., Silverberg, M.S., and Croitoru, K. (2014) Microbiome analysis – from technical advances to biological relevance. F1000Prime Reports 2014, 6:51 (doi:10.12703/P6-51). Downloaded from: : http://f1000.com/prime/reports/b/6/51.
- Smith, M.I., Turpin, W., Tyler, A.D., Silverberg, M.S., and Croitoru, K. (2014) Microbiome analysis – from technical advances to biological relevance. F1000Prime Reports 2014, 6:51 (doi:10.12703/P6-51). Downloaded from: : http://f1000.com/prime/reports/b/6/51.
- Smith, M.I., Turpin, W., Tyler, A.D., Silverberg, M.S., and Croitoru, K. (2014) Microbiome analysis – from technical advances to biological relevance. F1000Prime Reports 2014, 6:51 (doi:10.12703/P6-51). Downloaded from: : http://f1000.com/prime/reports/b/6/51.
- Smith, M.I., Turpin, W., Tyler, A.D., Silverberg, M.S., and Croitoru, K. (2014) Microbiome analysis – from technical advances to biological relevance. F1000Prime Reports 2014, 6:51 (doi:10.12703/P6-51). Downloaded from: : http://f1000.com/prime/reports/b/6/51
- Downloaded from: https://www.bing.com/images/search?view=detailV2&ccid=dbPKA3dM&id=3F381506D51B476F6CD0CC2B315C384CBE38F212&thid=OIP.dbPKA3dMyAnFjRZpay9o1gAAAA&mediaurl=http%3A%2F%2Funiverse-review.ca%2FI11-50-shotgun.jpg&exph=367&expw=367&q=Shotgun+DNA+Sequencing&simid=608042123925323991&ck=7631C710D9EC17F409F342861E4E129E&selectedindex=4&form=IRPRST&ajaxhist=0&vt=0&sim=11
- Smith, M.I., Turpin, W., Tyler, A.D., Silverberg, M.S., and Croitoru, K. (2014) Microbiome analysis – from technical advances to biological relevance. F1000Prime Reports 2014, 6:51 (doi:10.12703/P6-51). Downloaded from: : http://f1000.com/prime/reports/b/6/51
- Smith, M.I., Turpin, W., Tyler, A.D., Silverberg, M.S., and Croitoru, K. (2014) Microbiome analysis – from technical advances to biological relevance. F1000Prime Reports 2014, 6:51 (doi:10.12703/P6-51). Downloaded from: : http://f1000.com/prime/reports/b/6/51
- Downloaded from: https://www.bing.com/images/search?view=detailV2&ccid=s8lltPzp&id=04176E51799D7D07A0D71205525FADAA52D7DFCA&thid=OIP.s8lltPzpyT2XQBT451OE_gHaKs&mediaurl=https%3a%2f%2fwww.intechopen.com%2fmedia%2fchapter%2f38870%2fmedia%2fi mage1.jpeg&exph=1621&expw=1122&q=transcriptomics+analysis&simid=608001884327119411&ck=E9E7188E7C05D592583FDEF0B8D1B356&selectedIndex=4&FORM=IRPRST&ajaxhist=0
- Smith, M.I., Turpin, W., Tyler, A.D., Silverberg, M.S., and Croitoru, K. (2014) Microbiome analysis – from technical advances to biological relevance. F1000Prime Reports 2014, 6:51 (doi:10.12703/P6-51). Downloaded from: : http://f1000.com/prime/reports/b/6/51
- Smith, M.I., Turpin, W., Tyler, A.D., Silverberg, M.S., and Croitoru, K. (2014) Microbiome analysis – from technical advances to biological relevance. F1000Prime Reports 2014, 6:51 (doi:10.12703/P6-51). Downloaded from: : http://f1000.com/prime/reports/b/6/51
- Downloaded from: https://en.wikipedia.org/wiki/Metabolomics#/media/File:Key_stages_of_a_metabolomics_study.png.
- Christey, P. (issued by Hall, H.) (2020) As microbiome studies surge, new tools are needed. R&D World. Downloaded from: https://www.rdworldonline.com/as-microbiome-studiessurge-new-tools-are-needed/
- Downloaded from: https://www.bing.com/images/search?view=detailV2&ccid=a9PtrtNB&id=62D0234E97A48A218198200D670A1634EFDFA72E&thid=OIP.a9PtrtNBZGIJBL6PgnzuwQHaFk&mediaurl=https%3A%2F%2Fimage.slidesharecdn.com%2Fintroductiontonextgenerationsequencingv2-110927082709-phpapp02%2F95%2Fintroduction-to-nextgeneration-sequencing-14-728.jpg%3Fcb%3D1317179320&exph=547&expw=728&q=next+generation+sequencing&simid=608006840644406303&ck=05303D2BC342FB5C489ADEFFB9476D4F&selectedindex=17&form=IRPRST&ajaxhist=0&vt=0&sim=11
- Christey, P. (issued by Hall, H.) (2020) As microbiome studies surge, new tools are needed. R&D World. Downloaded from: https://www.rdworldonline.com/as-microbiome-studiessurge-new-tools-are-needed/