Water is crucial to pharmaceutical operations and the design of a water system within manufacturing facilities must minimize the opportunity for microbiological contamination. The type of water system used will depend on the process, or at which stage of manufacture the API is at, but it is paramount that the water is not detrimental to the product at any stage. There are a number of purification techniques that can be incorporated into the systems to reduce the microbial count, but as important is the regular testing regime of the water, to ensure the results are both accurate and reliable.
There are many definitions of what constitutes a viable organism. They include one that is capable of living, able to live on its own, able to reproduce, or even simply able to carry out normal cellular functions. These definitions may work well for multicellular organisms, but for bacteria, the picture is not quite so simple. For example, some bacterial cells that cannot replicate on artificial media can still carry out normal cellular functions and may regain their ability to reproduce, and some bacteria are symbiotes that cannot live and reproduce on their own.
The ability to grow a bacterial culture is often used as shorthand for viability, but this is also too simplistic. Organisms that reproduce are clearly viable, but the ability to grow depends on many factors, including available nutrients, pH, osmotic conditions and temperature. While culture remains the ‘gold standard’, just because a bacterial strain cannot be cultured, it does not mean it is not viable. The bacteria may be present in an environment where they cannot grow, but once inside an animal host they become infectious. Culture-based detection techniques, by definition, do not detect viable but non-culturable (VBNC) bacteria, but what methods can be used instead, and how successful are they?
The most common technique applied to the detection of VBNCs is fluorescence microscopy. Yet the plate-counting technique can still be more sensitive on identical samples, which implies that somewhere in the sample preparation process for microscopy, some of the bacteria are being lost – it can be as much as four orders of magnitude – or the staining methodology is not as effective as it might be. Comparative analyses using the commonly used visualization techniques, acridine orange, other nucleic acid stains, and 5-cyano-2,3-ditolyl tetrazolium chloride (CTC, a respiratory marker), invariably give different results.
As an example of just how different and unpredictable the results can be, a comparison of enumeration via culture with direct microscopic counts was made on three separate samples of drinking water, each disinfected in three different ways. As can be seen in Table 1, chlorine-treated water gave positive samples via plate count, CTC and acridine orange, though the levels were variable, and generally lower on the plate count. In contrast, no bacteria whatsoever were detected for ozone or UV treated water via plate count, and none with ozone treatment with CTC. They were detected for UV treatment with CTC, and in both cases with acridine orange. This suggests that these markers are not very effective in discriminating between live and dead bacteria, particularly for UV disinfection. UV causes fractures in the bacterial DNA, so organisms are unable to replicate, but both microscopy visualization techniques imply that viable bacteria remain present.
Table 1. Performance of microscopic counts with drinking water

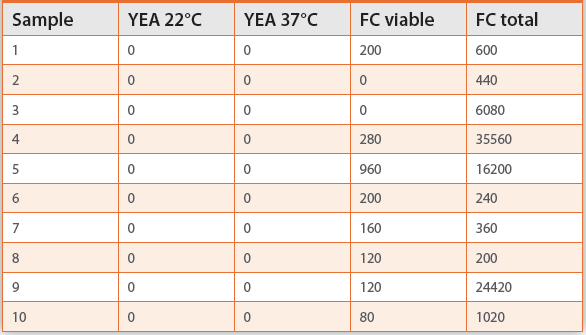
Flow cytometry is also used to check for the presence of viable bacteria in water samples, but is only suitable for samples with relatively high microbial counts such as potable water. Table 2 shows the results of a run of 10 test samples. In contrast to culture methods using yeast extract agar (YEA) which showed zero bacteria when incubated at either 22°C or 37°C, all those tested using flow cytometry indicated the presence of bacteria. In most cases, these included viable bacteria. This can be attributed to the fact that a single type of culture medium is used that is selective for bacteria of certain types at a specific temperature, rather than allowing all types of bacteria to grow. Flow cytometry is picking them all up, including those that will not grow on that particular medium.
Table 2. Flow cytometry data from drinking water
Another technique, solid phase scanning, finds some use in the pharmaceutical industry and other situations where the micro-biological quality of water is paramount. Here, in samples comparing two different media incubated at both 22°C and 37°C with the scanning method, as Table 3 shows, fewer bacterial cells were detected using yeast extract agar medium than with Reasoner´s 2A agar (R2A), a medium specifically designed for low nutrient environments. Significantly more bacteria were counted via solid phase scanning, however there is no consistent relationship between the number of organisms detected by culture or by laser scanning. Yeast extract agar is more commonly used, yet it picks up fewest bacteria in these tests. This, again, clearly shows that choice of test method has a significant impact on the results.
Table 3. Plate counts and laser scanning

A newer technique for the detection of viable bacteria relies on the polymerase chain reaction (PCR) in combination with intercalating dyes. PCR will detect anything that contains DNA, whether or not it is viable, and the dye introduces a degree of discrimination. If a cell is damaged, its membrane becomes non-selective in terms of what it allows to pass across it. Intercalating dyes are, typically, excluded from living cells, but will enter damaged ones, where they intercalate between the DNA bases, preventing the PCR from taking place, and providing a point of discrimination between live and dead cells.
As an example, 40 samples treated in different ways were each run for the presence of Pseudomonas aeruginosa using two different primer sets, one with an amplicon of approximately 200 base pairs, and a longer one with approximately 850 base pairs. With no treatment or culture, the results in Table 4 show there is no difference – every sample was positive. But with chlorine treatment, while most with the smaller amplicon were still positive, only 15 were with the larger one. Those treated with ozone or UV gave 13 and 17 positives respectively with the smaller amplicon, and 3 and 4 with the larger. The larger the amplicon, the more accurate the test is likely to be, as intercalation is more likely to occur. Intercalation is, essentially, a random process, and the larger the amplicon, the more likely it is to hit the DNA and prevent amplification. Yet it still remains imperfect, as ozone and UV treatment ought to kill all bacteria in the sample, and even with the 868bp amplicon, some positives remain.
Table 4. Detection on P.aeruginosa using PMA-PCR

One of the main factors governing the choice of procedure is the concentration of cells within the sample. Sample sizes for flow cytometry, for example, are of the order of 100μl, whereas with solid phase laser scanning samples that are liters in size can be run, albeit with some degree of difficulty, because of the filtration step. It is thus important to choose based on the number of cells that might be expected to be present, and the desired limit of detection.
It is also important to remember that there is no such thing as a nonselective culture procedure: one way or another, all culture methods are selective. Bacteria require conditions that are suitable for their growth, and the optimal nutrients, pH, oxygen levels and temperature differ from one bacterium to the next. For example, if the tests are being run on a product that’s designed for human consumption, detecting those bacteria that grow at body temperature will be of most interest. However, if spoilage is more of an issue, then testing for what remains viable at low temperature is more important.
In the pharmaceutical industry, it is common to culture using Tryptic Soy Agar (TSA) as the medium at 37°C, but in general this detects far less than R2A at 22°C, or yeast extract agar at either temperature. Yet the range of bacteria that will grow at body temperature is somewhat different to those that will grow under refrigeration conditions and cause spoilage on storage. Culturing at 37°C with TSA not only detects few bacteria, it also does not detect those that might cause problems with the product.
It is therefore important to choose the culture method appropriately. Overall, the majority of waterborne bacteria prefer lower temperatures, and the proportion of bacteria recovered increases with decreasing nutrient levels. Different combinations of nutrients and temperature select for different groups of organisms.
Low Nutrient Media
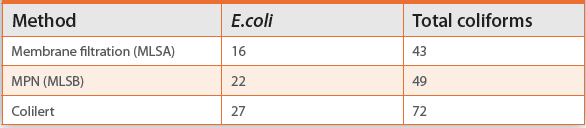
The use of liquid or solid media makes a big difference when detecting most bacteria, including E. coli and coliforms. In the comparison test whose results are shown in Table 5, samples were prepared and tested three ways – membrane filtration onto a solid medium, a liquid enrichment method with the same medium, and Colilert®, which is a low nutrient liquid medium. More organisms were detected using liquid enrichment than the solid medium, despite the fact that it was the same medium in both cases. Additionally, Colilert picks up significantly more of both E. coli and total coliforms. Clearly, a low nutrient medium is required for better detection.
Table 5. Recovery of E.coli and total coliforms
Similarly, with P. aeruginosa, the analogous low nutrient Pseudalert® system picks up many more positive samples than via either membrane filtration (the technique most commonly used for P. aeruginosa) or liquid enrichment, as indicated by the results in Table 6. Yet membrane filtration remains the primary technique used for water analysis, whether in the mains water supply, or in the pharmaceutical or food industry. A liquid medium with low nutrient levels puts the bacteria under less stress, making them more likely to grow and be detected.
Table 6. Recovery of Pseudomonas aeruginosa (142 samples)

Heterotrophic plate counts, or HPC, give a total count for viable bacteria, and again the choice of medium matters. Typically the media used are either TSA, YEA or R2A which all have very different nutrient levels. Using a similar low nutrient, liquid-based system can avoid at least some of these issues. The simple HPC for Quanti-Tray® test procedure allows 100ml test samples to be run, with growth detected by the cleavage of fluorochromes. Enumeration is a simple case of counting positive wells, with no interference.
In a comparison test between techniques, 44 samples were run at each of 22°C and 37°C, on two different media, the richer TSA that is commonly used in pharmaceutical water testing and the marginally less nutrient-rich YEA, and also low nutrient HPC for Quanti-Tray. Again, as shown in Table 7, HPC for Quanti-Tray gives higher counts. When YEA is compared with YEB, its liquid medium counterpart, and Quanti-Tray, the liquid version picks up a comparable number to Quanti- Tray.
Table 7. Comparison of liquid and solid media techniques for HPC (44 samples)

Liquid media in general are more likely to detect bacteria in water samples than solid media. Yet solid media still predominate, despite the fact that bacteria do not grow as well on them. It is thus advisable to use a liquid medium of some form if at all possible. The reasons for this are unclear, but – importantly in the pharma sector – if the bacteria have inadvertently been subjected to a sub-lethal dose of disinfectant because of an inappropriate cleaning procedure or disinfection strategy, using a form of test that is more likely to pinpoint problems is extremely advantageous.
It is known – but again, not fully understood - that if the organisms have been injured, such as via that sub-lethal disinfectant dose, then the benefits of liquid are further enhanced. When a liquid medium is used, it reaches the incubation temperature more slowly and, ideally, the sample would be incubated at the same temperature as it was when the sample was taken as it stresses bacteria less. However, there is a balance between the temperature that is used and the reason why the test is being done: if the major concern is to detect pathogenic bugs, the incubation temperature is likely to be higher.
Solid media also introduce a level of physical shock that might cause damage that is not present for liquid samples, and they are exposed instantly to 20% oxygen in the atmosphere, something which they are not used to if they have been living in the water supply. In liquid at 20°C, the oxygen level is likely to be closer to 9 or 10ppm. Oxygen can be toxic to bacteria, even if it is required for growth. It is the same with nutrients: too high a concentration can cause damage.
In most cases, it is best to carry out water sample tests in liquid rather than on solid media, preferably at low temperature, and with low levels of nutrients. The tests themselves should be carried out at the conditions that are optimized for the precise bacterium that is being sought. Most of the studies that have been done indicate that these conditions are likely to detect more of the viable bacteria that are present, even when compared to some of the modern analytical techniques that are presented as ideal for detecting viable non-culturable bacteria.
Author Biography
Professor Colin Fricker is managing director of CRF Consulting an international consulting firm specializing in microbiology of water, which based in the UK. Professor Fricker obtained his Ph.D. in microbiology from the university of Reading in 1987. In 1986, he was awarded the WH Pierce Prize for outstanding contributions to microbiology. He is an adjunct professor in the department of environmental sciences at Queens University, Kingston, Ontario, Canada. Professor Fricker has over thirty years experience in environmental microbiology, has edited five books, and published more than seventy peer reviewed publications.