School of Biomolecular and Biomedical Science, Conway Institute
Abstract
Heptahelical G protein-coupled receptors (GPCR) comprise a large family of integral membrane proteins involved in a wide array of cell signaling pathways. For high resolution structural studies of these receptors, multi-milligram quantities of pure and structurally unperturbed proteins are required. Purification of recombinant GPCRs typically involves their solubilization into detergent micelles followed by chromatographic purification. Because of relatively low expression levels of these recombinant receptors, it is challenging to design an efficient strategy for selective and efficient purification with high yield.
Here, we describe a recently introduced purification system employing a high affinity molecular switch based on fragment complementation, with a calcium dependent capture and EDTA mediated chelation elution. This technique was successfully applied to the purification of the recombinant cannabinoid receptor CB2, a promising target for the development of drugs for inflammation, immunological disorders and pain. It is feasible that similar strategies can be successfully employed for expression and purification of other membrane protein targets.
Introduction
G protein-coupled receptors (GPCR) comprise a large family of integral membrane proteins involved in a wide array of cell signaling pathways. The high resolution structural studies require large quantities of pure, homogenous and stable proteins. Purification of GPCRs typically requires solubilization of these receptors from cell membranes into detergent micelles, followed by one or more steps of affinity purification. This is notoriously challenging, because of the relatively low expression levels of GPCR in membranes and their instability in detergents. In recent years, significant improvements have been made in designing efficient protocols for expression of GPCRs in different cell types, their stabilization, and purification by affinity chromatography. 1-5
Human cannabinoid receptor CB2 is primarily expressed in cells of immune origin, and is an attractive target for the development of drugs for management of inflammation, immunological disorders and pain.6-8 Structural studies can provide critical contributions to the rational design of novel, specific drugs targeting this receptor.
Purification of a recombinant GPCR typically involves one or more steps of affinity chromatography, relying on engineered tag(s) fused to the expressed protein. Poly-histidine (immobilized-metal affinity chromatography, IMAC) and FLAG tags have been successfully used. 3, 9-15
Subscribe to our e-Newsletters
Stay up to date with the latest news, articles, and events. Plus, get special offers
from American Pharmaceutical Review – all delivered right to your inbox! Sign up now!
However, IMAC is not always efficient for purification of GPCRs, especially for those targets that are expressed at low levels, because of the low affinity of interaction between the His-tag and the resin in detergent containing buffers. The antibody-epitope interaction exploited by such systems as FLAG-tag/ FLAG resin may produce a high purity of the target protein, even in the presence of detergents.3,16 However, the application of FLAG resin for preparation of multi-milligram quantities of GPCR is expensive, especially in the case of commercial preparation of affinity resins. Several other affinity tag/matrix pairs have been described in the literature, but very few of them have been applied successfully to preparative-scale production of GPCR.17
When choosing the tag/affinity matrix pair for purification of membrane proteins for structural studies, the following parameters should be considered:
- High affinity in the presence of detergents (ideally in the picomolar or low nanomolar range)
- Ability to elute the captured protein in mild buffer conditions
- High binding capacity
- Ability to regenerate/re-use the matrix
- Low cost
- Small size of the affinity tag
Currently, there only a few examples of commercial kits that satisfy these requirements. One feasible alternative to the antibody/epitope interaction is a Strep-tag/StrepTactin system that relies on interaction between a small eight amino acid-long epitope (Strep-tag and a modified Streptavidin (StrepTactin). The recently introduced modification of the system StrepTactin XT affords pico- to nano-molar affinities between the Strep-tag and a resin, even in the presence of detergents.18 This approach has been successfully applied to purification of both soluble and membrane receptors.19 The limitation of the system is that the protein captured on the resin is eluted with high concentrations of biotin, and the elution kinetics are quite slow.
Because of its amphiphilic properties, biotin incorporates readily into detergent micelles, which may interfere with downstream applications of the purified protein.
An alternative strategy utilized in this work relies on a calcium dependent molecular switch based on the high affinity fragment complementation between the EF1 and EF2 domains of the Ca2+ -binding protein calbindin D9k.20-23 The high affinity binding of the two EF domains is based on interaction between hydrophobic core residues located in the α helices and loops of these fragments (Figure 1 A). The interaction between these domains can be broken by simply chelating the Ca2+ ions resulting in the loss of the helical secondary structure. Therefore, the elution step for EF-based purification of the recombinant protein can be performed at very mild conditions, at physiological pH, with addition of EDTA (Figure 1 B).
We tested the application of such Ca2+-dependent complementation in the purification of cannabinoid receptor CB2.25 The procedure takes advantage of the binding of calcium ions by the EF1 epitope fused to the recombinant receptor, and the EF2 affinity ligand coupled to the agarose matrix. In the presence of calcium, the helix-loop-helix motif on both EF1 and EF2 fragments is restored, facilitating a high-affinity binding of the target receptor to the resin.
Several constructs with the EF1 domain and other expression partners fused in various positions relative to the CB2 receptor were expressed in E. coli cells (Figure 2 A). The sequence of the DNA fragment encoding the 43 amino acid-long EF1 tag was optimized for codon usage of E. coli cells. Confirming our earlier observations for expression of CB2 receptor in bacterial cells,26 we determined that the presence of maltose binding protein (MBP) of E. coli as the N-terminal fusion partner was required for expression of CB2, and for its accumulation in bacterial cell membranes (Figure 2 B). When expressed with its signal sequence intact, the MBP is transported to the periplasmic space of E. coli, and is believed to act as a chaperone directing the downstream part of the fusion (CB2) to the cytoplasmic membranes.26 The periplasmic location also facilitates the formation of a disulfide bridge in the extracellular loop 2 of CB2, which is essential for stability of this receptor.27
Consequently, only the fusion constructs that harbor the N-terminal MBP fused to CB2 were expressed in E. coli cells; the construct CB2-351 (comprising the EF1-CB2 without an N-terminal MBP) did not appear to express as determined by Western blot with anti-CB2 antibody (Figure 2 B). Another significant observation is that the location of EF1 tag upstream of CB2 appears to have a negative effect on the expression levels of this fusion protein (construct 352). The location of EF1 tag at the C-terminus of the fusion (constructs 353, 354) did not have any negative effects on expression of these proteins.
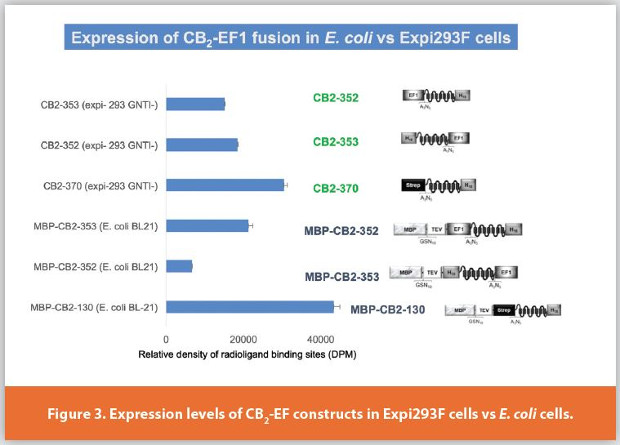
We further examined the expression of EF1-CB2 fusion proteins in other expression hosts besides E. coli. Figure 3 depicts the results of the expression of these constructs in suspension culture of Expi293F HEK cells. In these mammalian cells the constructs were expressed without the MBP fusion partner. The sequences of the 43 amino acid- EF1 tag, CB2, and a small linker were optimized for expression in mammalian cells. The expression levels of both CB2-352 (EF1 in the N-terminal position) and CB2-353 (EF1 in the C-terminal position) were approximately equivalent. Both constructs express at levels slightly lower than the control construct CB2-370 that harbors a twin-Strep-tag (22 amino acids including linker sequence) upstream of CB2. The expression levels of MBP-CB2-130 fusion construct in E. coli (normalized per milligram of membrane protein) were the highest among the construct and expression hosts tested. These findings underscore the importantance of the nature, size and the relative position of the affinity tag on the expression levels of the target protein. Naturally, the effects of the affinity tags on expression levels of the target protein also depend on a host cell. Our data show a potential utility of the EF1 tag for expression of integral membrane GPCR not only in a bacterial but also in a mammalian host.
The functionality of the recombinant receptor can be assessed by a ligand binding assay (typically, using a radioisotope-labeled ligand) and by measuring the activation of G protein on the receptor in response to ligand binding.27,28 Both assays are typically performed on preparations of cell membranes expressing the recombinant receptor. Figure 4A shows the saturation binding of the tritium-labeled high affinity agonist CP-55,940 on the membranes expressing construct CB2-354. The binding of the ligand is specific, with the Kd~ 1.4 nM, similar to the high affinity binding of this synthetic cannabinoid ligand to other EF1-CB2 expression constructs.25
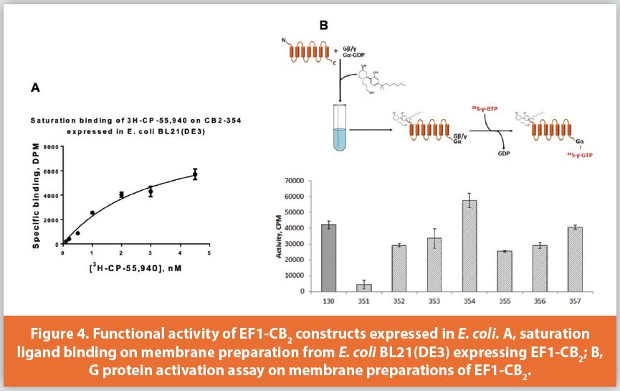
Another informative functional test quantifies the rates of activation of G protein on CB2 receptor in response to cannabinoid agonist binding. The assay can be performed with either bacterial membranes expressing CB2 protein or with purified receptor reconstituted into liposomes. Briefly, the receptor preparation is incubated with saturating concentrations of the activating ligand and is supplemented with purified subunits of G protein (in this case, Gαi1 and Gβ1γ2), in the presence of GDP. The reaction is initiated by the addition of radiolabeled analog of GTP, 35S-γ-GTP. This nucleotide analog is poorly hydrolyzible, and upon G protein activation on the receptor it tightly binds to the Gαi subunit. Thus, the complex of the labeled GTP with G protein accumulates for the duration of the reaction and provides a quantitative assessment of the amount of functional CB2 receptor. Figure 4B illustrates this concept by comparing membranes expressing different CB2 constructs. It reveals that the N-terminal (relative to CB2) location of EF1 tag is detrimental for the expression, while the C-terminal location of the tag results in higher expression levels. Furthermore, the placement of a small thioredoxin epitope between the C-terminus of CB2 and EF1 tag has a positive effect on expression levels of construct CB2-354. To summarize, these data confirm the correct folding and full functionality of EF1-CB2 constructs expressed in E. coli.
The purification of CB2 receptor performed using the EF1-EF2 complementation strategy with affinity resin prepared by thiol coupling of the EF2-GGC peptide to SulfoLink coupling agarose resin as described earlier.25 The recombinant fusion protein is solubilized from cell membranes in detergent micelles comprised of zwitterionic detergent CHAPS and non-ionic n-dodecyl-β-Dmaltopyranoside (DDM). For stabilization of the receptor, cholesteryl hemiscuccinate (CHS) and a high affinity ligand (CP-55,940) are added, and all purification buffers are supplemented with 15-30% (v/v) glycerol.
All steps of purification are performed at 4°C or on ice to preserve the correct fold of the recombinant protein.
The detergent extracts with solubilized receptor are mixed with EF2-agarose, and the binding is typically performed as a batch purification overnight, in the presence of Ca2+. The batch mode helps to maximize the binding capacity of the resin, which can be as high as 0.5 mg of protein per mL of resin.

The resin is then washed extensively with Ca2+ -containing buffers to ensure that the high affinity interaction between EF1 and EF2 epitopes is maintained. No significant loss of the captured protein is detected during this step. The elution is performed by the addition of EDTA that chelates Ca2+ ions. The protein bound to the affinity matrix is released gradually; typically, 4-5 column volumes of EDTA-containing elution buffer are necessary to complete the process (Fig. 5). Both the C-terminal location of the EF1 tag (constructs CB2-353, CB2-354) and mid-fusion location (construct CB2-352) are suitable for purification using this strategy, although in the case of mid-fusion EF1 tag, the binding capacity of the resin appears to decrease, possibly due to steric hindrance effects from the large MBP fusion partner. Also, in the case of construct CB2-352 some spontaneous cleavage of the fusion construct can occur during purification that results in the removal of MBP and release of the EF1-CB2. Importantly, a significant purity of the protein preparation can be attained after just one chromatography step, with an enrichment factor for the EF1-CB2 protein of at least 1000-fold.
Both the capture and elution steps are performed at neutral pH values. The mild condition of the purification preserve the functional structure of the receptor, as confirmed by activity tests of the purified products as described earlier.25
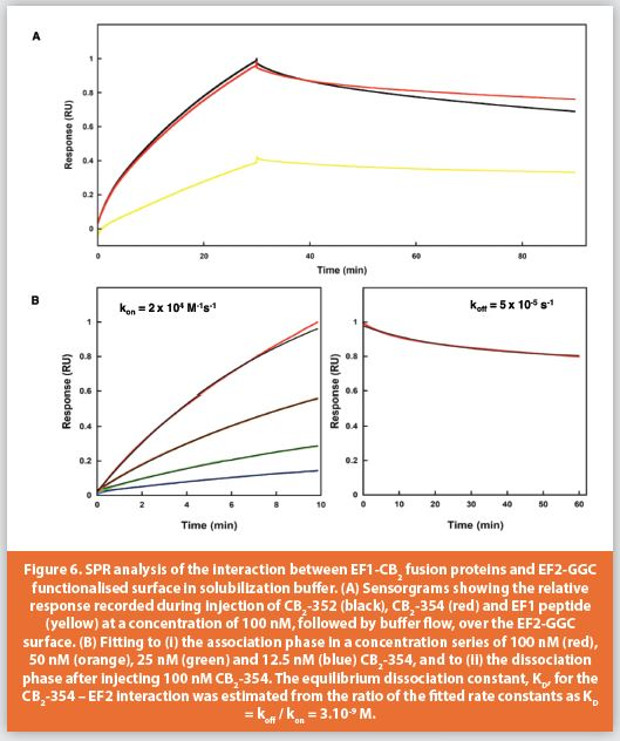
We next determined the binding kinetics between EF1-CB2 proteins and immobilized EF2-GGC in the detergent and glycerol rich solubilization buffer used to purify EF1-CB2 at 25, 10 and 4 degrees Celsius. EF2-GGC that was thiol coupled to the carboxymethyl dextran surface of a BiaCore CM5 sensor chip and purified EF1-CB2-352 and EF1-CB2-354 fusions were injected over the surface for 30 minutes followed by buffer flow for 60 minutes. EF1 wild type peptide was injected as a positive control (Figure 6A).25 The equilibrium dissociation constant, KD, for the CB2-354 – EF2 interaction was estimated from the ratio of the fi tted rate constants as KD = koff / kon = 3.10-9 M (Figure 6B) and 5.10-9 M for EF1-CB2-352, that had an apparently faster off rate than that of EF1-CB2-354, which while not significant, may be due to the N terminal location of the EF1-tag in this fusion. The maintenance of the high affinity of interaction between EF1 and EF2 sequences in this buffer at low temperature, with the simple method of elution, highlights the potential to optimize this system further in the capture, purification and analysis of functional integral membrane proteins.
To summarize, the use of EF1 tag for recombinant production of CB2 and affinity purification on EF2 matrix facilitates the isolation of the fully functional protein in just one chromatography step. The yield of the recombinant protein may be further improved by optimizing the expression conditions and purification parameters. The technology of preparation of the recombinant CB2 can be adapted to expression and purification of other GPCR and recombinant receptors and open a path for a meaningful biophysical characterization of these proteins.
Acknowledgements
This work was supported by the Intramural Research Program of the NIAAA, NIH and the Science Foundation Ireland Investigator Project Grant 12-IP-1620. Authors gratefully acknowledge the contribution by Dr. N.N. Mhurchu and Gavin McGauran (University College Dublin), Professor Sara Linse (Lund University), valuable technical assistance of Mr. K. Hines and Ms. L. Zoubak and continuous support by Dr. K. Gawrisch (NIAAA, NIH). Expression in Expi293 cells was performed by the group of Dr. J. Zmuda (ThermoFisher Scientific).
References
- Das, M., Du, Y., Mortensen, J. S., Hariharan, P., Lee, H. S., Byrne, B., Loland, C. J., Guan, L., Kobilka, B. K., and Chae, P. S. (2018) Rationally Engineered Tandem Facial Amphiphiles for Improved Membrane Protein Stabilization Efficacy, Chembiochem 19, 2225-2232.
- Weis, W. I., and Kobilka, B. K. (2018) The Molecular Basis of G Protein-Coupled Receptor Activation, Annu Rev Biochem 87, 897-919.
- Rasmussen, S. G. F., Choi, H.-J., Fung, J. J., Pardon, E., Casarosa, P., Chae, P. S., DeVree, B. T., Rosenbaum, D. M., Thian, F. S., Kobilka, T. S., Schnapp, A., Konetzki, I., Sunahara, R. K., Gellman, S. H., Pautsch, A., Steyaert, J., Weis, W. I., and Kobilka, B. K. (2011) Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor, Nature 469, 175-180.
- Yeliseev, A. A., and Vukoti, K. (2011) Expression of G protein-coupled receptors, In Production of membrane proteins (Robinson, A. S., Ed.), pp 219-248, Wiley-VCH, Weinheinm, Germany.
- Park, S. H., Das, B. B., Casagrande, F., Tian, Y., Nothnagel, H. J., Chu, M., Kiefer, H., Maier, K., De Angelis, A. A., Marassi, F. M., and Opella, S. J. (2012) Structure of the chemokine receptor CXCR1 in phospholipid bilayers, Nature 491, 779-783.
- Graham, E. S., Ashton, J. C., and Glass, M. (2009) Cannabinoid receptors: A brief history and “what’s hot”, Frontiers in Bioscience-Landmark 14, 944-957.
- Munro, S., Thomas, K. L., and Abushaar, M. (1993) Molecular characterization of a peripheral receptor for cannabinoids, Nature 365, 61-65.
- Pacher, P., and Mechoulam, R. (2011) Is lipid signaling through cannabinoid 2 receptors part of a protective system?, Progress in Lipid Research 50, 193-211.
- Britton, Z., Young, C., Can, O., McNeely, P., Naranjo, A., and Robinson, A. S. (2011) Membrane protein expression in Saccharomyces cerevisiae, In Production of membrane proteins (Robinson, A. S., Ed.), pp 37-74, Wiley-VCH, Weinheim, Germany.
- Casagrande, F., Maier, K., Kiefer, H., Opella, S. J., and Park, S. H. (2011) Expression and purification of G-protein-coupled receptors for nuclear magnetic resonance structural studies, In Production of membrane proteins (Robinson, A. S., Ed.), pp 297-316, Wiley-VCH, Weinheim, Germany.
- Koehl, A., Hu, H. L., Maeda, S., Zhang, Y., Qu, Q. H., Paggi, J. M., Latorraca, N. R., Hilger, D., Dawson, R., Matile, H., Schertler, G. F. X., Granier, S., Weis, W. I., Dror, R. O., Manglik, A., Skiniotis, G., and Kobilka, B. K. (2018) Structure of the mu-opioid receptor-G(i) protein complex, Nature 558, 547-+.
- Mancia, F., and Hendrickson, W. A. (2007) Expression of recombinant G-protein coupled receptors for structural biology, Molecular bioSystems 3, 723-734.
- Rosenbaum, D. M., Zhang, C., Lyons, J. A., Holl, R., Aragao, D., Arlow, D. H., Rasmussen, S. G. F., Choi, H.-J., DeVree, B. T., Sunahara, R. K., Chae, P. S., Gellman, S. H., Dror, R. O., Shaw, D. E., Weis, W. I., Caffrey, M., Gmeiner, P., and Kobilka, B. K. (2011) Structure and function of an irreversible agonist-beta(2) adrenoceptor complex, Nature 469, 236-240.
- Sun, B., Bachhawat, P., Chu, M. L., Wood, M., Ceska, T., Sands, Z. A., Mercier, J., Lebon, F., Kobilka, T. S., and Kobilka, B. K. (2017) Crystal structure of the adenosine A2A receptor bound to an antagonist reveals a potential allosteric pocket, Proc Natl Acad Sci U S A 114, 2066-2071.
- Thal, D. M., Sun, B., Feng, D., Nawaratne, V., Leach, K., Felder, C. C., Bures, M. G., Evans, D. A., Weis, W. I., Bachhawat, P., Kobilka, T. S., Sexton, P. M., Kobilka, B. K., and Christopoulos, A. (2016) Crystal structures of the M1 and M4 muscarinic acetylcholine receptors, Nature 531, 335-340.
- Jaakola, V.-P., Griffith, M. T., Hanson, M. A., Cherezov, V., Chien, E. Y. T., Lane, J. R., Ijzerman, A. P., and Stevens, R. C. (2008) The 2.6 Angstrom crystal structure of a human A(2A) adenosine receptor bound to an antagonist, Science 322, 1211-1217.
- Kimple, M. E., Brill, A. L., and Pasker, R. L. (2013) Overview of affinity tags for protein purification, Curr Protoc Protein Sci 73, Unit 9 9.
- Schmidt, T. G. M., Batz, L., Bonet, L., Carl, U., Holzapfel, G., Kiem, K., Matulewicz, K., Niermeier, D., Schuchardt, I., and Stanar, K. (2013) Development of the Twin-Strep-tag (R) and its application for purification of recombinant proteins from cell culture supernatants, Protein Expression and Purification 92, 54-61.
- Yeliseev, A., Zoubak, L., and Schmidt, T. G. M. (2017) Application of Strep-Tactin XT for affinity purification of Twin-Strep-tagged CB2, a G protein-coupled cannabinoid receptor, Protein Expression and Purification 131, 109-118.
- Lewit-Bentley, A., and Rety, S. (2000) EF-hand calcium-binding proteins, Curr Opin Struc Biol 10, 637-643.
- Yamniuk, A. P., Gifford, J. L., Linse, S., and Vogel, H. J. (2008) Effects of metal-binding loop mutations on ligand binding to calcium- and integrin-binding protein 1. Evolution of the EF-hand, Biochemistry-Us 47, 1696-1707.
- Berggard, T., Julenius, K., Ogard, A., Drakenberg, T., and Linse, S. (2001) Fragment complementation studies of protein stabilization by hydrophobic core residues, Biochemistry-Us 40, 1257-1264.
- Linse, S., Voorhies, M., Norstrom, E., and Schultz, D. A. (2000) An EF-hand phage display study of calmodulin subdomain pairing, J Mol Biol 296, 473-486.
- Svensson, L. A., Thulin, E., and Forsen, S. (1992) Proline cis-trans isomers in calbindin D9k observed by X-ray crystallography, J Mol Biol 223, 601-606.
- Mhurchu, N. N., Zoubak, L., McGauran, G., Linse, S., Yeliseev, A., and O’Connell, D. J. (2018) Simplifying G Protein-Coupled Receptor Isolation with a Calcium-Dependent Fragment Complementation Affinity System, Biochemistry-Us 57, 4383-4390.
- Krepkiy, D., Wong, K., Gawrisch, K., and Yeliseev, A. (2006) Bacterial expression of functional, biotinylated peripheral cannabinoid receptor CB2, Protein Expression and Purification 49, 60-70.
- Yeliseev, A. A. (2013) Methods for Recombinant Expression and Functional Characterization of Human Cannabinoid Receptor CB2, Comput Struct Biotec 6.
- Vukoti, K., Kimura, T., Macke, L., Gawrisch, K., and Yeliseev, A. (2012) Stabilization of functional recombinant cannabinoid receptor CB2 in detergent micelles and lipid bilayers, PLOS One 7, e46290.