Introduction
Microbial identification places an important role in pharmaceutical processing. Microbial identification can be defined as “microbial characterization by a limited spectrum of tests pre-chosen and appropriate to the problem being studied” [1]. Characterizing a microorganism can provide important information as to its origin and potential impact in relation to a product or in relation to the environment in which it was isolated. Microbiologists should understand the quantity and the types of microorganisms present in pharmaceutical ingredients, water for pharmaceutical use, in-process materials, final product and the manufacturing environment.
Classically microbial identification was undertaken using staining techniques and various agars and tests aimed at differentiating one probable species from another. This process was advanced during the 1970s through the advent of the API (analytical profile index) test strip, consisting of a series of militarized biochemical substrates contained within ampoules [2]. Since the 1990s, a series of phenotypic methods became available for the QC Microbiology laboratory in a format that was relatively easy to use and relatively affordable. The 2000s saw an equivalent range of genotypic methods introduced, designed for the routine processing of pharmaceutical process samples [3].
The genotype–phenotype distinction between identification systems is drawn in genetics. Genotype refers to an organism’s full hereditary information (representing its exact genetic makeup); whereas phenotype describes an organism’s actual observed properties, such as morphology, reaction to different chemicals, or behavior. Phenotypic methods allow the microbiologist to identify microbial species to the genus and sometimes to the species level based on a relatively small number of observations and tests. These are primarily growth-dependent methods, and identification must begin with a pure culture. In contrast to the examination of the phenotypic characteristic of a microorganism, genotypic techniques study the microbial genome itself [4].
Examples of phenotypic automated systems include automated identification and susceptibility tests as well as automated incubatorreaders [5, 6], Fourier-Transform Infrared (FTIR) spectroscopy [7], and Matrix-Assisted Laser Desorption Ionization–Time of Flight (MALDITOF) mass spectroscopy [8, 9], which study the spectra produced by different microorganisms. An alternative method is the use of cellular fatty acid (FA) composition. With this method fatty acids are extracted from the cell cultures and then the patterns of fatty acid esters are determined by gas chromatography [10]. Phenotypic methods rely on specified culture media and incubation conditions.
In terms of genotypic methods, these include riboprinting. This system is an automated Southern Blot apparatus using labeled ssDNA probe from the 16sRNA codon [11]. A second group of methods are PCR-based. The technology sequences the first 500 base pairs of the unknown microorganism’s 16s ribosomal RNA codon. The technology involves amplification of the 16S codon by PCR, followed by automated sequencing [12]. Genotypic methods are more robust than phenotypic methods because they are not affected by cultural conditions. The technologies are, however, more technically challenging for microbiologists without training in molecular biology.
There are a number of factors to consider when choosing between systems. According to Sutton, these will include costs (including an assessment of both validation costs, consumable costs and operating costs); throughput, in terms of samples processed per day or per week; the time-to-result; the labor requirements; the amount of facility space needed; and compatibility with existing systems like LIMS [13]. To this, Fung adds acceptability by the scientific community; simplicity of operation, including training requirements, and reagents; and technical services provided by the vendor [14].
Whilst the pharmaceutical microbiologist now faces a somewhat bewildering array of different test systems, once the selection has been made it is important, from a GMP perspective, that the system is validated. For this, both the United States Pharmacopeia and European Pharmacopeia offer some guidance. In general, the two approaches share more similarities than differences. There are, however, some aspects which are not in agreement. These are important to understand for those working for global pharmaceutical organizations, where inspections from different national regulators may occur. The purpose of this paper is to present and discuss the most important of these differences.
Pharmacopeia Guidance
Pharmacopeia guidance for the validation of microbial identification systems is provided by the United States Pharmacopeia (USP) and by the European Pharmacopeia (EP). The USP guidance is contained within a specific chapter: “Microbial Characterization, Identification and Strain Typing” <1113>, which became official from December 1, 2012 [15], whereas the EP guidance is contained within its general chapter on rapid and alternative microbiological methods (Ph. Eur. 5.1.6 “Alternative Methods for Control of Microbiological Quality”), which was introduced in 2006 and is currently undergoing revision [16].
USP Chapter <1113>
There are three important parts of USP. The first part addresses the general workflow for microbial identification, covering aspects like the use of pure cultures (required for phenotypic methods, rather than genotypic methods which are usually unaffected by cultural conditions) and methods for primary screening or pre-screening (such as the Gram-stain and spore stain). It is within this section that the main phenotypic and genotypic identification methods are outlined. In the second part, the verification of microbial identification methods is outlined. It is noticeable here that the term ‘verification’ is used, which contrasts to the EP’s use of the term ‘validation’. The final part of the chapter consists of a glossary of terms relating to microbial taxonomy and identification.
With the verification requirements, the USP offers three approaches that can be taken. These are:
- Parallel testing with approximately fifty microbial isolates using an existing system
- The testing of twelve to fifteen representative stock cultures of commonly isolates species (ensuring that these are of a broad enough range to cover the majority of the instruments test array), here type strains should ideally be used
- Confirming that 20-50 microbial identifications, including 15-20 different species, agree with the results of a reference laboratory testing split sample.
Whichever of the three approaches is selected, the USP recommends that appropriate QC organisms, recommended by the supplier and appearing in the official chapters of the compendia, should be included. This would include microorganisms suitable for the qualification of culture media and in relation to the sterility test and tests for microbial enumeration and specified microorganisms. The USP then proceeds to outline the most important verification tests. These are:
- Accuracy, which is expressed as a percentage of the number of correct results divided by the number of obtained results, multiplied by 100
- Reproducibility, which is similarly expressed as a percentage. Here the number of correct results in agreement is divided by the total number of results multiplied by 100
This means, for each instrument verification, two index scores are produced.
Other measurements to consider are sensitivity, specificity and the positive/negative predictive value (which is applicable for the detection of specific microorganisms).
EP Chapter 5.1.6
The European Pharmacopeia has a different structure to that of the USP. There are two key parts of the chapter relating to microbial identification. These are, first, an assessment of common microbial identification methods, which considers the principles of measurement, the critical aspects of operation, and potential uses. Secondly, the chapter provides advice about general validation, primary method validation (which is generally undertaken by the vendor), and the validation for the actual intended use (which is undertaken by the user to demonstrate method applicability).
With the assessment of methods, the EP chapter divides these into classical identification methods (methods for defining morphological characteristics, such as the Gram-stain, and techniques for defining biochemical characterization, which would include the API strip and equivalent methods) and into alternative methods (which includes everything else including rapid methods).
With ‘alternative methods’, several common identification methods are described (Table 1).
Table 1. Table displaying types of microbial identifi cation methods described in the European Pharmacopeia
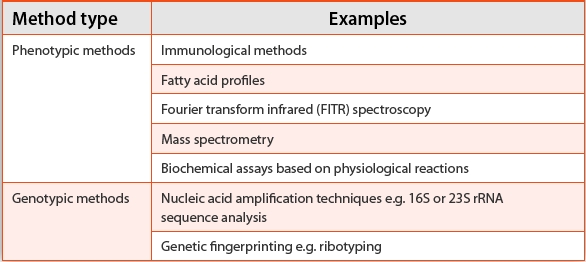
The European Pharmacopeia provides guidance on system validation and recommends a three-step method. This consists of:
Step 1: General Validation
With step 1, the pharmacopeia states that equipment must be first qualified through the standard qualification processes: Design Qualification, Installation Qualification, Operational Qualification, and Performance Qualification.
At this stage, the pharmacopeia also advises that sequence databases should have adequate coverage in relation to the microorganisms of interest to the user (although it offers no further guidance on how to make such an evaluation, presumably the user will need crossreference the typical cleanroom and water system microbiota against the instrument’s database).
Step 2: Primary Method Validation
With the second stage, the pharmacopeia outlines what is expected of the vendor of the instruments. This includes describing accurately the principle of detection and method of use. With the method, the supplier must state the materials and equipment required. The pharmacopeia also recommends that any instrument marketed for a pharmaceutical facility must have been published in at least one peer-reviewed journal.
Step 3: Validation for the Actual Intended Use
This stage outlines what the user of the instrument is expected to undertake in order to qualify the instrument within the laboratory. This consists of assessing three areas: accuracy, precision and robustness. Whilst no acceptance criteria are given, they are defi ned, as set out, with explanation, in Table 2.
Table 2. Key validation criteria from the European Pharmacopeia for the validation of automated microbial identifi cation methods.

Notably, the European Pharmacopeia advises against attempting to verify one microbial identification method against another, which is in contrast to the fi rst of the USP’s three approaches for method verification. The EP’s reluctance to recommend comparison is due to diff erent methods varying in terms of their respective abilities to identify microorganisms from different products and samples and due to different systems working in different ways and having different databases.
Comparing the Pharmacopeia
The guidance for microbial identification in the two pharmacopeia are generally well aligned. The differences are outlined in Tables 3 and 4. Important areas of agreement relate to the types of technologies available, their descriptions and advice in the selection of a method. Furthermore, it is apparent that neither pharmacopeia recommends the use of anything other than compendia or type cultures for verifying the identification outcome against the instrument’s database. The use of environmental isolates (or ‘wildtypes’) is not referred to. Presumably this is because environmental isolates are too variable and would not allow for inter-laboratory assessments. However, the USP and EP approaches differ most notably with terminology (verification or validation) and with the numbers and types of approaches designed to achieve the adoption of an automated identification method into the laboratory.
Table 3. Selection of identification methods, comparison of USP and EP.

Table 4. Selection of verification / validation approaches, comparison of USP and EP.

With the method verification, the point of agreement is the use of type cultures (the USP second option and the EP’s reference to accuracy and precision). This is an obvious starting point for the qualification of a rapid microbial identification method where a need to meet the requirements of both pharmacopeia is required. In contrast, the main disagreement is with whether an attempt should be made to compare one system with another, with the USP presenting this as an acceptable approach and the EP specifically cautioning against this approach. With the USP’s third option, the use of a reference laboratory, the EP makes no mention.
The use of a reference laboratory raises a number of potentially difficult questions, including which reference laboratory to choose?; how is the reference laboratory accredited?; how can the reference laboratory be shown to be accurate?; how can it be decided that the identification system used by the reference laboratory is accurate?; and, what would be the likely outcome should the reference laboratory use a very different identification system with a database composed of several different species? Some care is required before selecting this option.
Conclusion
This article has outlined the important of microbial identifi cation and characterization for the pharmaceutical industry and has proceeded to present the applicable guidance for the selection of different identification systems and approaches for their verification (or, alternatively, their ‘validation’) as set out in the United States and European Pharmacopeia.
Through the exercise of comparing and contrasting the two pharmacopeia chapters, it is clear that they are broadly aligned. When one chapter offers information not covered in detail by the other, the microbiologist would do well to read both and to ensure that guidance is taken from both compendia. Here the USP’s coverage of morphological and biochemical screening (especially the need for a pure culture for phenotypic methods) should be considered alongside the EP’s reference to equipment qualifi cation steps.
Where the pharmacopeia diff er most is, depending which path is followed, the core verifi cation or validation. To steer a path between them would involve the use of suitable type cultures and undertaking an assessment for accuracy, precision and robustness.
Whether these differences are reduced once the revised EP 5.1.6 chapter is published remains to be seen. In the meantime, selecting the points of comparison and where they complement one another, remains a best practice strategy.
Author Biography
Tim Sandle, Ph.D., is the Head of Microbiology at the Bio Products Laboratory. Dr. Sandle is responsible for a range of microbiological tests, batch review, microbiological investigation and policy development. In addition, Dr. Sandle is an honorary consultant with the University of Manchester and is a tutor for the university’s pharmaceutical microbiology MSc course, and he runs an on-line pharmaceutical microbiology site (www. pharmamicro.com). Dr. Sandle is a chartered biologist and holds a first class honors degree in Applied Biology; a Master degree in education; and a Ph.D. in microbiology.
Dr. Sandle can be reached at: [email protected]
References
- Stackebrandt,E. (1989) Taxonomic considerations Prochloron: publications and perspective: 65-69.
- PDA. 2000. Evaluation, Validation and Implementation of New Microbiological Testing Methods, Technical Report No. 33, PDA, Vol. 54, No. 3, May/June 2000, Supplement TR33
- O’Hara, C.M.,Weinstein, M.P., and Miller, J.M. (2003): Manual and Automated Systems for Detection and Identifi cation of Microorganisms, ASM Manual of Clinical Microbiology, 8”’Edition
- Sutton, S. V. W. and Cundell, A. M. (2004): Microbial Identifi cation in the Pharmaceutical Industry, Pharmacopeial Forum, Vol. 35, No.5, pp.1884 – 1894
- Gilardi, G. L. (1990). Identifi cation of glucose-nonfermenting gram negative rods. North General Hospital, New York
- Funke, G., and Funke-Kissling, P. (2004). Evaluation of the New VITEK 2 Card for Identifi cation of Clinically Relevant Gram-Negative Rods, J Clin Microbiol. 42(9):4067- 4071, 2004
- Maquelin, K., Kirschner, C., Choo-Smith, L. P., van den Braak, N.,
- Endtz, H. P., Naumann, D. & Puppels, G. J. (2002). Identifi cation Of Medically Relevant Microorganisms By Vibrational Spectroscopy, J Microbiol Meth. 51:255-271
- Fox, A. (2006). Mass Spectrometry for Species or Strain Identifi cation after Culture or without Culture: Past, Present, and Future, J Clin Microbiol. 44(8):2677-2680
- Mazzeo, M.F. Sorrentino, S., Gaita, M., Cacace, G., Di Stasio, M. Facchiano, A., Comi, G., Malorni, A., and Siciliano, R.A. (2006). Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry for the Discrimination of Food-Borne Microorganisms, Appl Environ Microbiol. 72(2):1180-1189
- Eerola, E and O-P Lehtonen (1988). Optimal Data Processing Procedure for Automatic Bacterial Identifi cation by Gas-Liquid Chromatography of Cellular Fatty Acids, J Clin Microbiol. 9:1745-1753
- Brisse, S. Verduin, C.M., Milatovic, D., Fluit, A., Verhoef, J., Laevens, S., Vandamm, P., Tummler, B., Verbrugh, H.A., and van Belkum, A. (2000). Distinguishing Species of the Burkholderia cepacia Complex and Burkholderia gladioli by Automated Ribotyping, J Clin Microbiol. 38(5):1876–1884, 2000
- Fontana, C., Favaro, M., Pelliccioni, M., Pistoia, E.S. and Favalli, C. (2005) Use of the MicroSeq 500 16S rRNA Gene-Based Sequencing for Identifi cation of Bacterial Isolates that Commercial Automated Systems Failed to Identify Correctly, J Clin Microbiol. 43(2):615-619
- Sutton, S.V. (2011). Qualifi cation of a Microbial Identifi cation System, Journal of Validation Technology, 17 (4): 46-49
- Fung D.Y.C. (2002). Rapid Methods and Automation in Microbiology. Comprehensive Rev. in Food Science and Food Safety 1:3-22
- USP. (2010) <1113> Microbial Characterization, Identifi cation and Strain Typing. Pharmacopeial Forum. 36(6):1675–1688
- EP. (2006). Chapter 5.1.6 Alternative Methods for Control of Microbiological Quality, European Pharmacopeia, European Directorate for the Quality of Medicines & HealthCare, Brussels, Belgium.